Decision trees and random forests
MELODEM data workshop
Wake Forest University School of Medicine
Overview
Decision trees
Growing trees
Leaf nodes
Random Forests
Out-of-bag predictions
Variable importance

Data were collected and made available by Dr. Kristen Gorman and the Palmer Station, a member of the Long Term Ecological Research Network.
Decision trees
Growing decision trees
Decision trees are grown by recursively splitting a set of training data (Breiman 2017).
Gini impurity
For classification, “Gini impurity” measures split quality:
\[G = 1 - \sum_{i = 1}^{K} P(i)^2\]
\(K\) is no. of classes, \(P(i)\) is the probability of class \(i\).
First split: right node
Consider splitting at flipper length of 206.5: Right node:
First split: left node
Consider splitting at flipper length of 206.5: Left node:
First split: total impurity
Consider splitting at flipper length of 206.5: Impurity:
Your turn
Was this the best possible split? Let’s find out.
Open
classwork/02-trees_and_forests.qmdComplete exercise 1.
Reminder: pink sticky note if you’d like help, blue sticky note when you are finished.
Hint: the answer is 0.492340868530637
05:00
Regression trees
Impurity is based on variance:
\[\sum_{i=1}^n \frac{(y_i - \bar{y})^2}{n-1}\]
\(\bar{y}\): mean of \(y_i\) in the node.
\(n\): no. of observations in the node.
Survival trees
Log-rank stat measures split quality (Ishwaran et al. 2008):
\[\frac{ \sum_{j=1}^J O_{j } - E_{j}}{\sqrt{\sum_{j=1}^J V_{j}}}\] \(O\) is observed and \(E\) expected events in the node at time \(j\). \(V\) is variance.
Keep growing?
Now we have two potential datasets, or nodes in the tree, that we can split.
Do we keep going or stop?
Stopping conditions
We may stop tree growth if the node has:
Obs <
min_obsCases <
min_casesImpurity <
min_impurityDepth =
max_depth
Your turn
Suppose
min_obs= 10min_cases= 2min_impurity= 0.2max_depth= 3
Which node(s) can we split?
Split the left node
The left node can be split.
The right node cannot, since impurity on the right is <
min_impurity
Finished growing
The left node can be split.
After splitting the left node, all nodes have impurity <
min_impurity.If no more nodes to grow, convert partitioned sets into leaf nodes
Leaves?
What is a leaf node?
A leaf node is a terminal node in the tree.
Predictions for new data are stored in leaf nodes.
mutate(
penguins,
node = case_when(
flipper_length_mm >= 206.5 ~ 'leaf_1',
bill_length_mm >= 43.35 ~ 'leaf_2',
TRUE ~ 'leaf_3'
)
) %>%
group_by(node) %>% count(species) %>%
mutate(n = n / sum(n)) %>%
pivot_wider(values_from = n, names_from = species,
values_fill = 0)# A tibble: 3 × 4
# Groups: node [3]
node Adelie Chinstrap Gentoo
<chr> <dbl> <dbl> <dbl>
1 leaf_1 0.016 0.04 0.944
2 leaf_2 0.0635 0.921 0.0159
3 leaf_3 0.966 0.0345 0 Your turn
Fit your own decision tree to the penguins data.
Open
classwork/02-trees_and_forests.qmdComplete Exercise 2
05:00
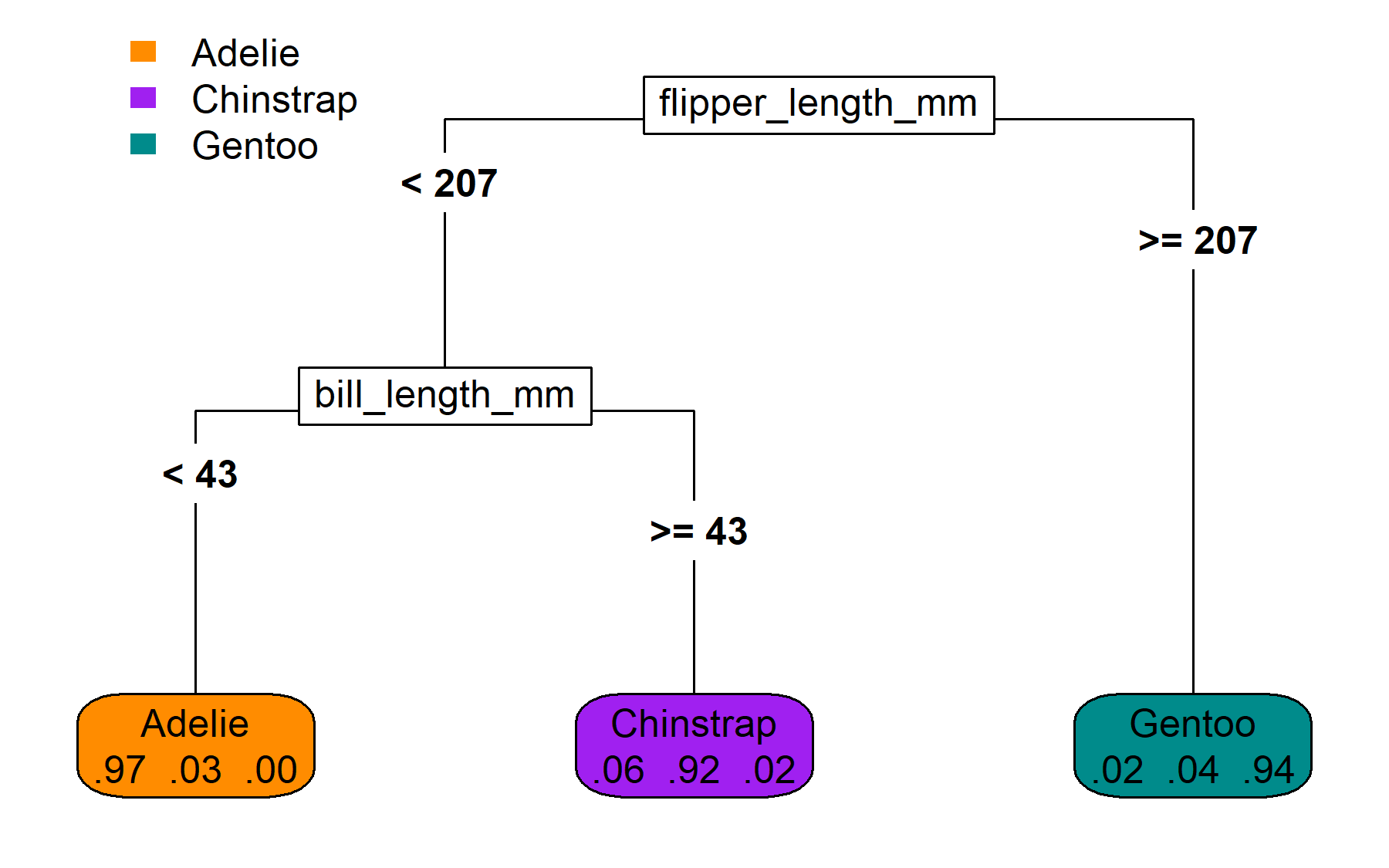
From partition to flowchart
The same partitions, visualized as a binary tree.
As a reminder, here is our ‘hand-made’ leaf data
| Adelie | Chinstrap | Gentoo | |
|---|---|---|---|
| leaf_1 | 0.02 | 0.04 | 0.94 |
| leaf_2 | 0.06 | 0.92 | 0.02 |
| leaf_3 | 0.97 | 0.03 | 0.00 |
Leaves for other types of trees
For regression, leaves store the mean value of the outcome.
For survival, leaves store:
cumulative hazard function
survival function
summary score (e.g., no. of expected events)
Planting seeds for causal trees
Suppose we have outcome \(Y\) and treatment \(W\in\{0, 1\}\).
\[\text{Define }\tau = E[Y|W=1] - E[Y|W=0],\]
If \(\tau\) can be estimated conditional on a person’s characteristics, then a decision tree could be grown using \(\hat\tau_1 \mid x_1, \ldots, \hat\tau_n \mid x_n\) as an outcome. That tree could predict who benefits from treatment and explain why.
But how do we get \(\hat\tau_i \mid x_i\)? More on this later…
Random forests
Expert or committee?
If we have to make a yes/no decision, who should make it?
1 expert who is right 75% of the time
Majority rule by 5000 independent “weak experts”.
- Note: each weak expert is right 51% of the time.
Answer is weak experts!
Why committee?
If we have to make a yes/no decision, who should make it?
1 expert who is right 75% of the time
Majority rule by 5000 independent “weak experts”.
- Note: each weak expert is right 51% of the time.
Answer is weak experts!
Why? Weak expert majority is right ~92% of the time
Random forest recipe
The weak expert approach for decision trees (Breiman 2001):
Grow each tree with a random subset of the training data.
Evaluate a random subset of predictors when splitting data.
These random elements help make the trees more independent while keeping their prediction accuracy better than random guesses.
A single traditional tree is fine
A single randomized tree struggles
Five randomized trees do okay
100 randomized trees do great
500 randomized trees - no overfitting!
Random forest predictions
To get a prediction from the random forest with \(B\) trees,
predict the outcome with each tree
take the mean of the predictions
\[\hat{y}_i = \frac{1}{B}\sum_{b=1}^{B} \hat{y}_b(i)\]
Out-of-bag
Breiman (1996) discusses “out-of-bag” estimation.
‘bag’: the bootstrapped sample used to grow a tree
‘out-of-bag’: the observations that were not in the bootstrap sample
out-of-bag prediction error: nearly optimal estimate of the forest’s external prediction error
With conventional bootstrap sampling, each observation has about a 36.8% chance of being out-of-bag for each tree.
How to get out-of-bag predictions
Grow tree #1
Store its predictions and denominators
Now do tree #2
Accumulate predictions and denominator
Out-of-bag predictions
Final out-of-bag predictions:
\[\hat{y}_i^{\text{oob}} = \frac{1}{|B_i|}\sum_{b \in B_i}\hat{y}_b\]
- \(B_i\) is the set of trees where \(i\) is out of bag
Out-of-bag estimation plays a pivotal role in variable importance estimates and inference with causal random forests.
Variable importance
First compute out-of-bag prediction accuracy.
Here, classification accuracy is 96%
Variable importance
Next, permute the values of a given variable
See how one value of flipper length is permuted in the figure?
Variable importance
Next, permute the values of a given variable
Now they are all permuted, and out-of-bag classification accuracy is now 61%
Variable importance
Rinse and repeat for all variables.
For bill length, out-of-bag classification accuracy is reduced to 63%
Variable importance
Once all variables have been through this process:
\[\text{Variable importance} = \text{initial accuracy} - \text{permuted accuracy}\]
Flipper length: 0.96 - 0.61 = 0.35
Bill length: 0.96 - 0.63 = 0.33
\(\Rightarrow\) flippers more important than bills for species prediction.
Note: Details on limitations in Strobl et al. (2007)
Your turn
Complete exercise 3.
05:00
References
Slides available at https://bcjaeger.github.io/melodem-apoe4-het/