TODO: add description
scalib_hare(
scalib_object,
verbose = 0,
hare_penalty = NULL,
hare_max_dimension = NULL,
hare_prophaz = FALSE,
hare_additive = FALSE,
hare_linear_risk = FALSE,
hare_linear_time = FALSE,
hare_fit = NULL
)Arguments
- scalib_object
An object of class
scalib(see scalib).- verbose
(integer value) If 0, no output will be printed. If 1, some output will be printed. If 2, all output will be printed.
- hare_penalty
the parameter to be used in the AIC criterion. The method chooses the number of knots that minimizes
-2 * loglikelihood + penalty * (dimension). The default is to use penalty = log(samplesize) as in BIC. The effect of this parameter is summarized in summary.hare.- hare_max_dimension
maximum dimension (default is
6 * length(data)^0.2.- hare_prophaz
should the model selection be restricted to proportional hazards models?
- hare_additive
should the model selection be restricted to additive models?
- hare_linear_risk
(logical value) should the effect of risk be linear?
- hare_linear_time
(logical value) should the effect of time be linear?
- hare_fit
hare object. If
fitis specified, hare adds basis functions starting with those in fit.
Value
an object of class scalib
Examples
# packages
library(ggplot2)
library(tidyr)
library(dplyr)
#>
#> Attaching package: ‘dplyr’
#> The following objects are masked from ‘package:data.table’:
#>
#> between, first, last
#> The following objects are masked from ‘package:stats’:
#>
#> filter, lag
#> The following objects are masked from ‘package:base’:
#>
#> intersect, setdiff, setequal, union
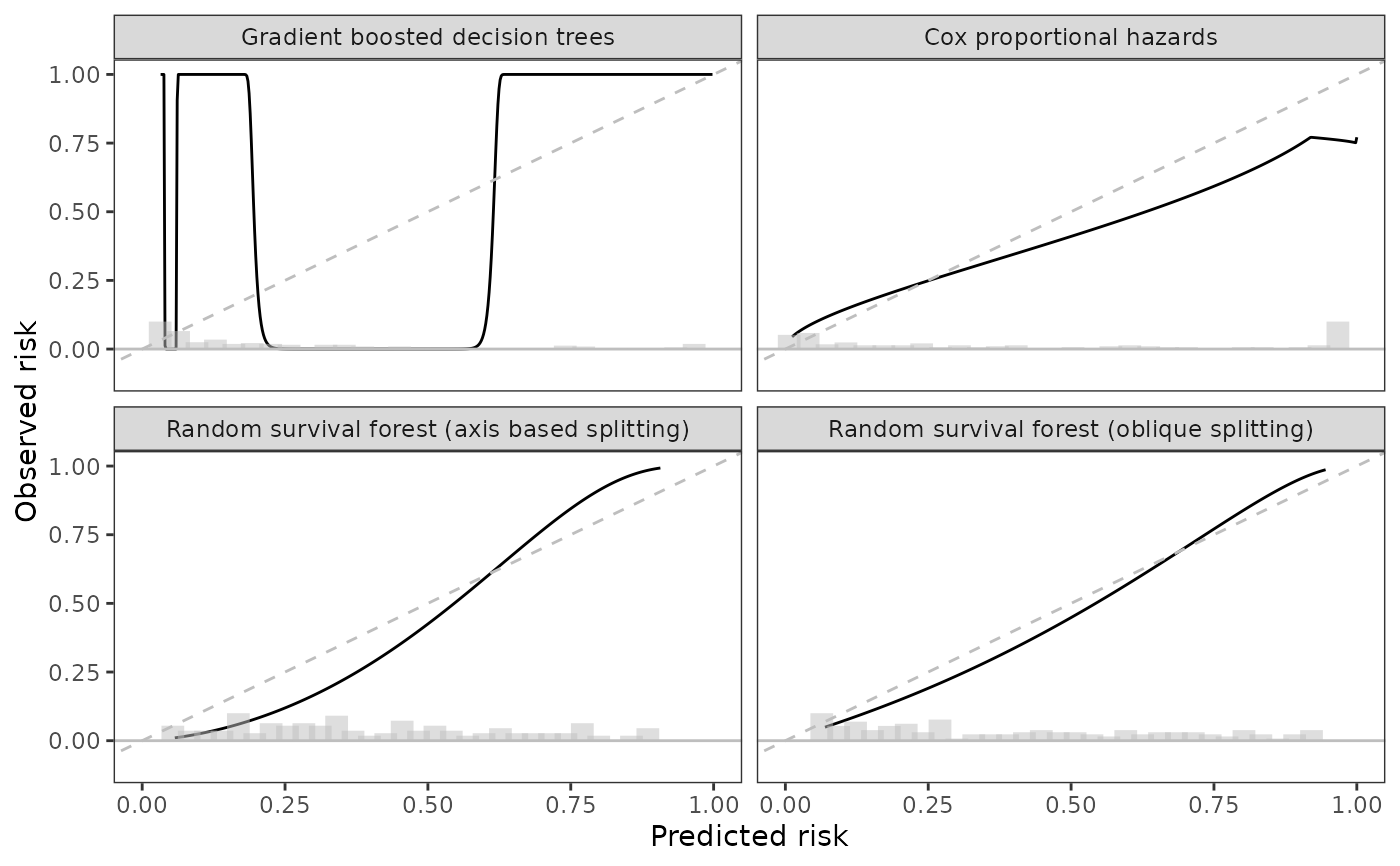
sc <- scalib(pred_risk = pbc_scalib$predrisk,
pred_horizon = 2500,
event_time = pbc_scalib$test$time,
event_status = pbc_scalib$test$status)
sc_hare <- scalib_hare(sc)
print(sc_hare)
#>
#> Survival calibration object with prediction horizon of 2500
#>
#> -- Input data ----------------------------------------------------------------
#>
#> event_time event_status prop_hazard rsf_axis gradient_booster rsf_oblique
#> <int> <num> <num> <num> <num> <num>
#> 1: 400 1 0.9990 0.9026 0.9351 0.9463
#> 2: 4500 0 0.4272 0.3072 0.0524 0.3680
#> 3: 1925 1 0.8286 0.4722 0.2342 0.5982
#> 4: 1832 0 0.0358 0.1474 0.0422 0.1460
#> 5: 2466 1 0.0392 0.1925 0.0568 0.1558
#> ---
#> 134: 1300 0 0.1509 0.1783 0.0629 0.1669
#> 135: 1293 0 0.1805 0.3466 0.1299 0.2416
#> 136: 1250 0 0.9823 0.4743 0.3254 0.5727
#> 137: 1230 0 0.0182 0.0589 0.0322 0.0637
#> 138: 1153 0 0.0718 0.1637 0.0527 0.1220
#>
#>
#> -- Output data -------------------------------------------------------------
#>
#> Key: <._id_.>
#> ._id_. hare_ici hare_e50 hare_e90 hare_emax hare_data_plot
#> <char> <num> <num> <num> <num> <list>
#> 1: gradient_booster 0.5104 0.4045 0.9358 0.969 <data.table[500x2]>
#> 2: prop_hazard 0.1008 0.0454 0.2404 0.248 <data.table[500x2]>
#> 3: rsf_axis 0.0999 0.1068 0.1335 0.836 <data.table[500x2]>
#> 4: rsf_oblique 0.0417 0.0449 0.0622 0.065 <data.table[500x2]>
#> 1 variable not shown: [hare_data_smry <list>]
#>
data_gg <- sc_hare$data_outputs |>
select(._id_., hare_data_plot) |>
unnest(hare_data_plot)
data_bins <- predrisk_bin_segments(sc_hare, bin_count = 30) |>
mutate(._id_.)
id_labels <- c(prop_hazard = "Cox proportional hazards",
rsf_axis = "Random survival forest (axis based splitting)",
gradient_booster = "Gradient boosted decision trees",
rsf_oblique = "Random survival forest (oblique splitting)")
ggplot(data_gg) +
aes(x = predicted, y = observed) +
geom_line() +
geom_abline(col = 'grey', linetype = 2) +
geom_hline(yintercept = 0, col = 'grey') +
theme_bw() +
theme(panel.grid = element_blank()) +
coord_cartesian(xlim = c(0, 1),
ylim = c(-0.1, 1)) +
geom_segment(data = data_bins,
size = 4,
alpha = 0.50,
col = 'grey',
mapping = aes(x = x,
y = y,
xend = xend,
yend = yend)) +
facet_wrap(~._id_., labeller = labeller(._id_. = id_labels)) +
labs(x = 'Predicted risk', y = 'Observed risk')