library(survival.calib)
library(survival)
library(riskRegression)
library(dplyr)
library(ggplot2)
knitr::opts_chunk$set(fig.width=7, fig.height=5)Overview
Data
For this example, we’ll use a standard dataset, survival::flchain, with some very light modifications.
# drop rows with missing values for simplicity
data_init <- na.omit(flchain)
data_init$chapter <- NULL
head(data_init)
#> age sex sample.yr kappa lambda flc.grp creatinine mgus futime death
#> 1 97 F 1997 5.70 4.860 10 1.7 0 85 1
#> 2 92 F 2000 0.87 0.683 1 0.9 0 1281 1
#> 3 94 F 1997 4.36 3.850 10 1.4 0 69 1
#> 4 92 F 1996 2.42 2.220 9 1.0 0 115 1
#> 5 93 F 1996 1.32 1.690 6 1.1 0 1039 1
#> 6 90 F 1997 2.01 1.860 9 1.0 0 1355 1We’ll also use a simple split of the data for training and testing.
Model fitting
Suppose we have two model specifications that we want to compare:
Model 1: use
trt,sex, andstageto predict risk for mortalityModel 2: use
age,biliandplateletto predict risk for mortality.
Code to fit these models to the training data is below:
Code to fit these models to the training data is below:
model_1 <- coxph(Surv(futime, death) ~ age + sex,
data = data_train,
x = TRUE)
model_2 <- update(model_1, . ~ . + lambda + kappa + creatinine)
# compute predicted risk at 2000 days post baseline
time_predict <- c(2000)
predrisk_1 = predictRisk(model_1, newdata = data_test, times = time_predict)
predrisk_2 = predictRisk(model_2, newdata = data_test, times = time_predict)
scal_2000 <- scalib(pred_risk = list(predrisk_1[,1],
predrisk_2[,1]),
pred_horizon = time_predict,
event_status = data_test$death,
event_time = data_test$futime) %>%
scalib_gnd() %>%
scalib_hare()Now let’s combine the data and create a graph showing both curves.
data_calslope <- bind_rows(scal_2000$data_outputs$hare_data_plot,
.id = 'model')
fig_cal_slopes <- ggplot(data_calslope) +
scale_y_continuous(limits = c(-0.2, 1.2),
breaks = seq(0, 1, by = 0.2),
labels = paste0(seq(0, 100, by = 20),"%")) +
aes(x = predicted, y = observed, color = model) +
geom_line() +
geom_abline(col = 'grey', linetype = 2, intercept = 0, slope = 1) +
scale_x_continuous(limits = c(0,1),
expand = c(0,0),
breaks = seq(0, 1, by = 0.2),
labels = paste0(seq(0, 100, by = 20),"%")) +
theme_bw() +
theme(panel.grid = element_blank())
fig_cal_slopes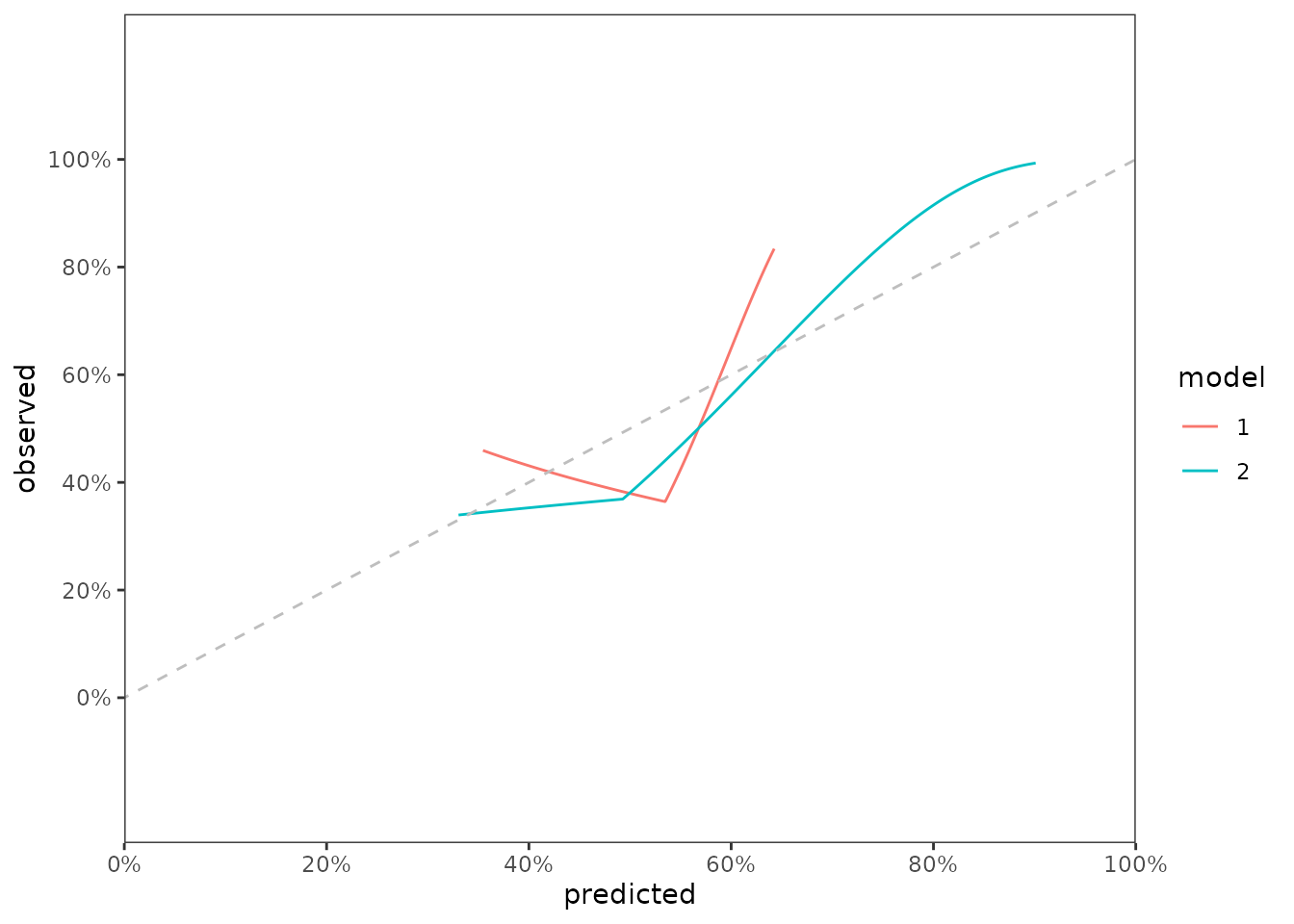
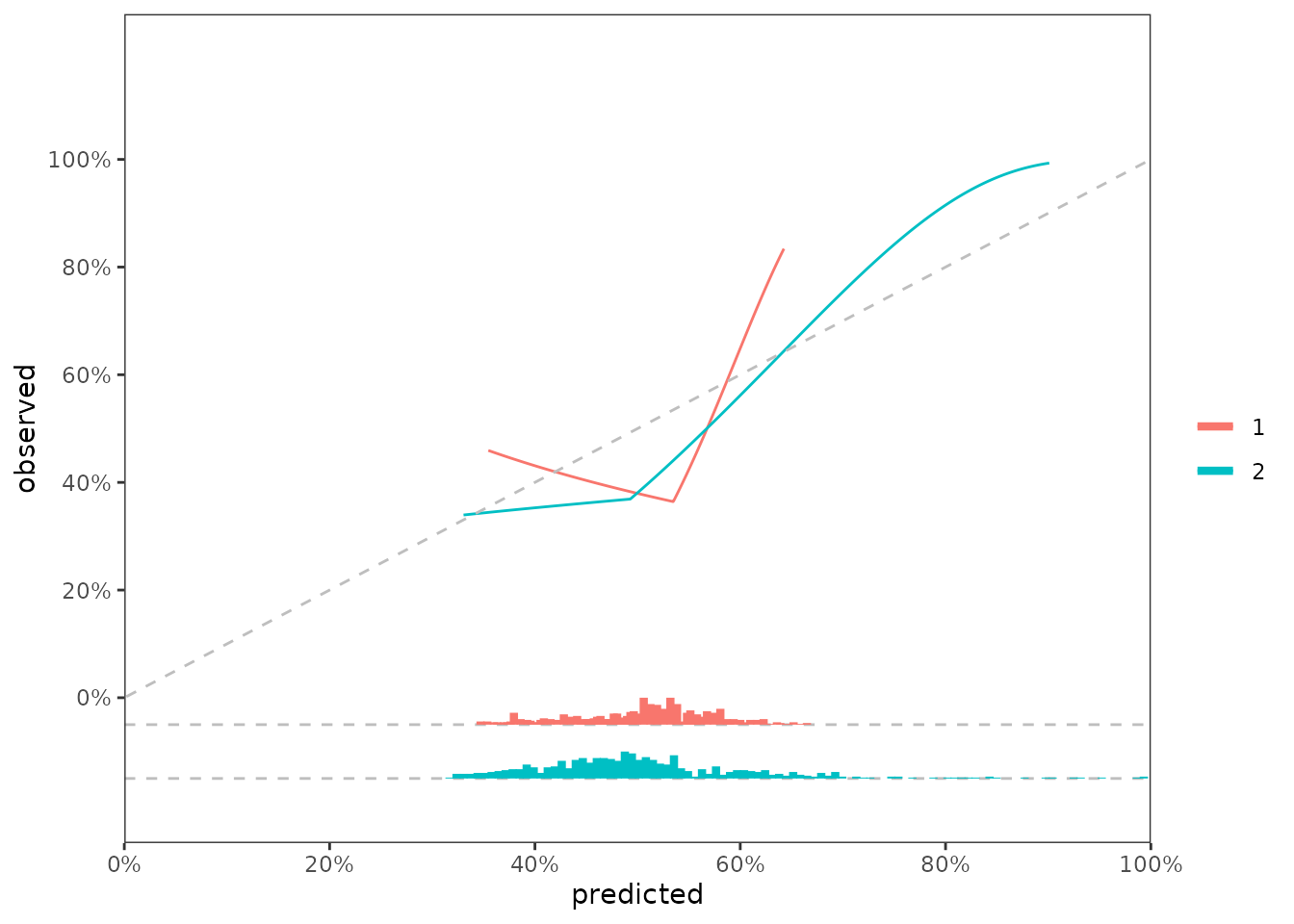
It’s standard practice to show the distribution of predicted risk underneath the calibration slope plots. For this visualization, we can use predrisk_bin_segments, which creates a dataframe containing all of the aesthetics required for geom_segment().
segment_1 <- predrisk_bin_segments(x = predrisk_1,
event_status = data_test$status,
event_time = data_test$time,
bin_yintercept = -0.05,
bin_length = 1/2)
segment_2 <- predrisk_bin_segments(x = predrisk_2,
event_status = data_test$status,
event_time = data_test$time,
bin_yintercept = -0.15,
bin_length = 1/2)
data_segment <- bind_rows(segment_1, segment_2,.id = 'model')
fig_cal_slopes +
geom_hline(yintercept = -0.05, linetype = 2, color = 'grey') +
geom_hline(yintercept = -0.15, linetype = 2, color = 'grey') +
geom_segment(data = data_segment,
inherit.aes = FALSE,
size = 1.5,
mapping = aes(x = x,
y = y,
color = model,
xend = xend,
yend = yend)) +
labs(color = '')